This worked example will walk you through the insertion of GELLO code on decision arms for the '? anaemia' decision in the GORD (Gastro-oesophageal reflux) example. A GLIF file is provided (GORD_start.GLIF) that is suitable to use; along with some test patient data in xml format.
Anaemia is a low haemoglobin (hemoglobin) level, and in the GORD guideline example represents a compelling reason for early endoscopy, as there may be a cancer of the esophagus or stomach causing the reflux symptom.
Open and unzip the attached folder. Open the GLIF file with the GLIF Editor.
It opens at the "top level" guideline. There are three sub-guidelines and they can be accessed by clicking through the guideline itself or by scrolling down on right hand side (RHS) of the menu bar:
Go to the "consider early endoscopy" sub-guideline.
Notice the decisions, as elsewhere are pink. They are not automated as yet. So we will do that for the anaemia decision.
It's good (for me anyway) to write GELLO for GLIF decisions against loaded patient data. Let's load some data. This is in xml format and you can look at the test data in notepad++ as well if you like.
Go back to the top level guideline then go File, Load xml Test data. Choose test.xml. Go back to the "consider early endoscopy" guideline.
Now click on the Allow modifications button ( count six in from the LHS of the top menu.)
Position the cursor on the "yes" arrow coming out of the "Anaemia?" decision such that you get a little hand with the finger pointing onto the arrow. Double click and go Edit:
.. to get to the GELLO editor screen:
The main panes are the middle workspace and the RHS Model Explorer, Data Explorer and Results Explorer.
Lets take a minute to look at the Model Explorer.
Unfold on System Packages. From the bottom up, GLIF_VMR models a representation of GLIF that accords with 'level B' described on page 7 of the document GLIF_TECH_SPEC_May_4_2004.pdf. So this is the model that understands where and what we can see and do with GELLO in Level A GLIF artefacts such as nodes/steps. Make particular note of the attributes required for a GLIFDecision Result as we use them later on. Above GLIF_VMR is HL7_v2_VMR_V1. This is a draft HL7 standard virtual medical record model that Medical Objects balloted successfully in 2013. Most of the world uses V2 messaging so we can build a CDS suitable model out of what people currently use and send back and forth.
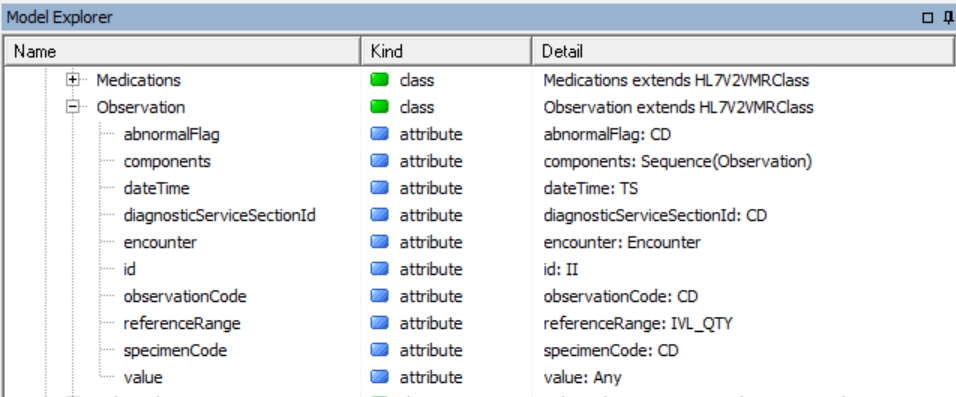
Notice some classes in there such as Allergy and Observation. These classes have attributes that use ISO 21090 data types such as CD and TS:
Fold all that up and the next level up is SNOMED CT. SNOMED CT is a large medical ontology looked after by IHTSDO.org. GELLO uses it especially for subsumption queries, with the "implies"method. We won't see this today but it is useful in the homework at the end ;-)
Above this is iso_21090_datatypes. If you look up the CD datatype in here you can see that is about coded values and that the implies method is understood by this datatype.
Ok lets go back to the middle workspace. Notice the editor puts the first line in for you. We are in a context of a GLIF Decision node.
Our first line of GELLO below this context statement will be:
1 + 1