- Created by Peter Scott, last modified on Aug 28, 2024
Introduction to the editor
Put the downloaded file mowgli_11.exe in a folder called 'Gello'' for example in C: drive. You can make a shortcut to it if you like, or pin it to the taskbar. Run it and you should have this:
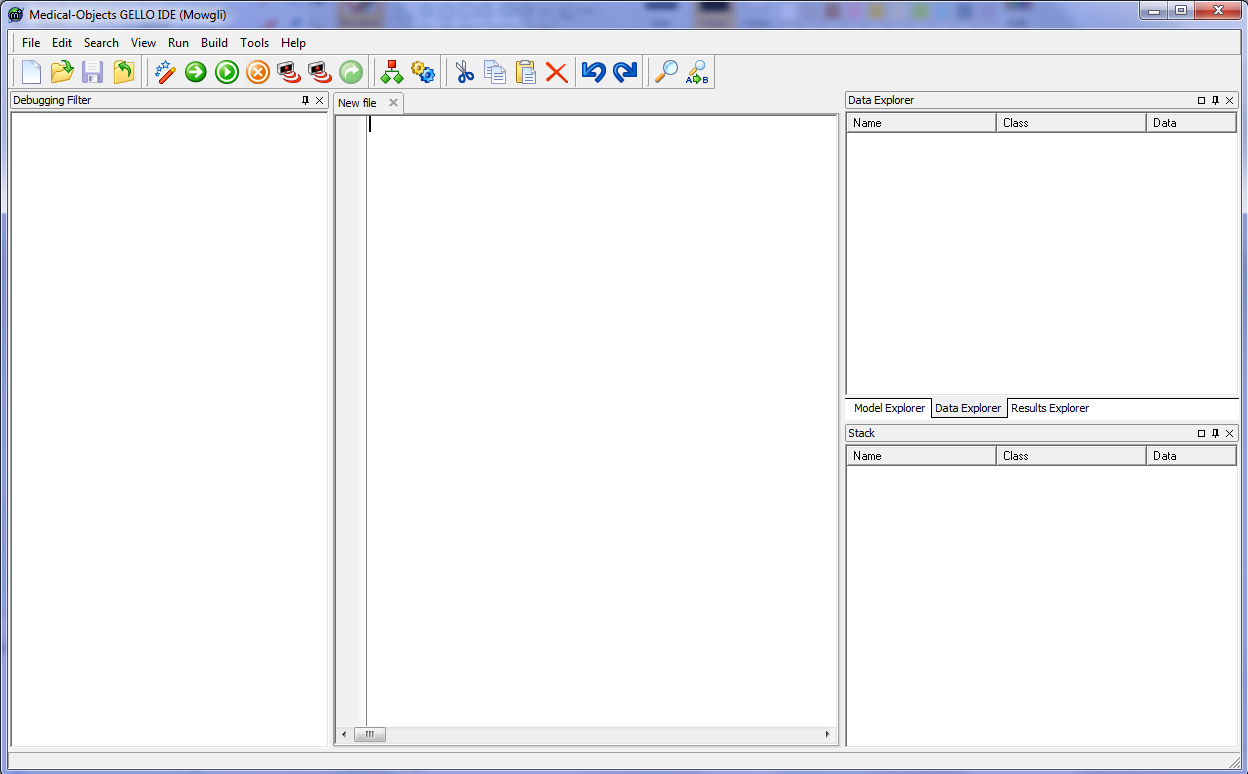
On the left hand side (LHS) there is a debugging filter which we won't use so minimise up that window by adjusting the pane size, and make some more room for the RHS panes as well:
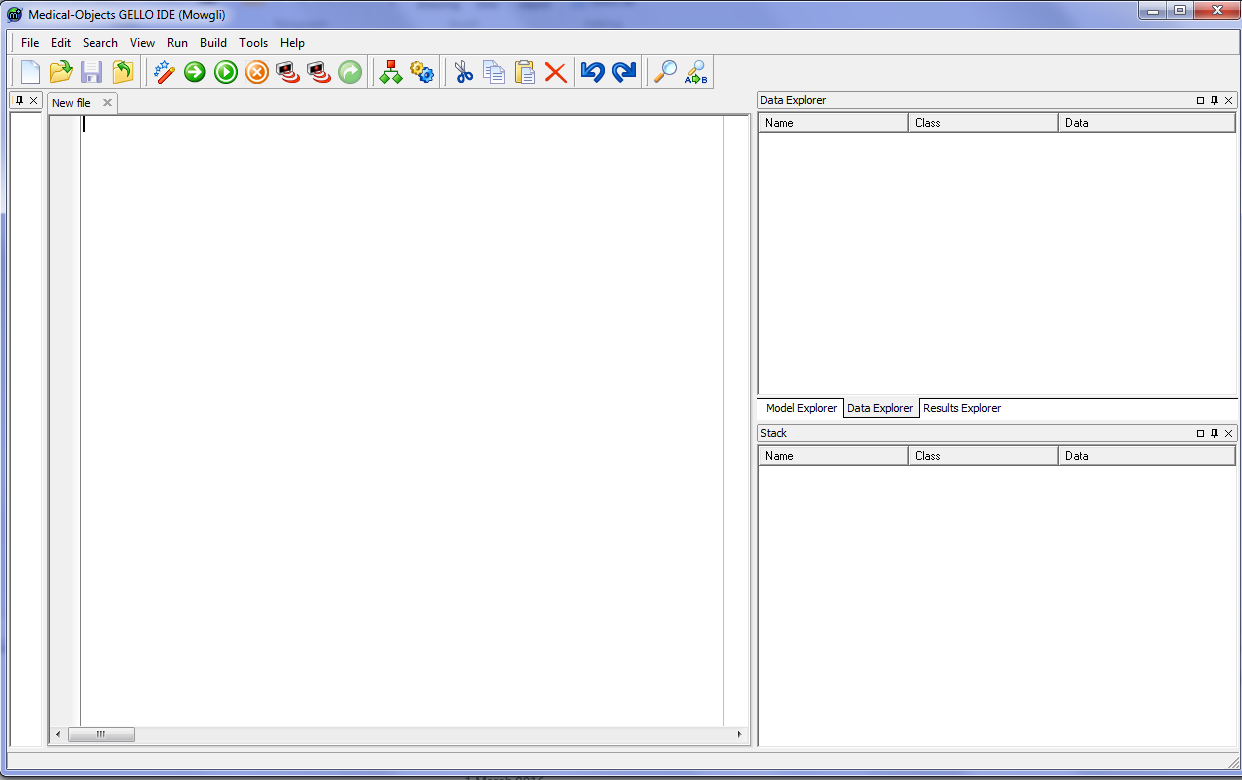
So, looking at the editor now we have a largish workspace area where we will write GELLO code, a top menu bar, and on the RHS, a Model Explorer, a Data Explorer and a Results Explorer (in the tabs of the top RHS pane), and a stack for debugging also. We won't use the Stack viewer very much however the pane above it is very important so give it some more vertical room.
GELLO is a query language for writing computer executable decision support in the health domain. It can access information out of a virtual medical record (VMR). A VMR is an information model that presents what is useful for clinical decision support out of an Electronic Health Record. So this will be things like patient age and gender, their past illnesses, operations, medications , allergies etc. GELLO also can perform logic against a reference terminology such as SNOMED CT which means it can do things like ask if the patient has a respiratory disease, without having to type in all possible codes for all possible respiratory diseases to find out.
Before we get started writing our first piece of code take a look at the Model Explorer tab on RHS. Unfold on System Packages:
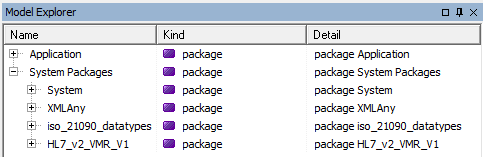
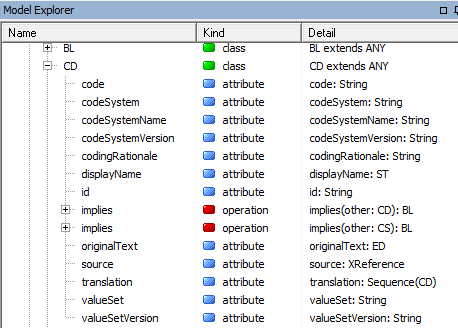
We'll take a look at the bottom two packages. These describe the ISO 21090 datatypes that GELLO uses (GELLO is strongly typed) and the HL7 v2 VMR which is the default VMR model - although other VMRs may be used. Unfold on iso_21090_datatypes, and scroll down and you can see a large number of types. We will use standard datatypes such as String, Real, Boolean (These are in fact in the System package further up in the last screenshot), and from iso_21090_datatypes - mostly we will use Concept Descriptor (CD), Physical Quantity (PQ), Timestamp (like a datetime) (TS) and the Sequence classes. Codes and code systems get used a lot in health IT and if you unfold the CD datatype class you can see the attributes and operations available to this type:
Now fold all that up and have a look at the HL7_v2_VMR_V1 package. If you are a clinician you will recognise this as a fairly comprehensive summary of clinical information for a patient. Unfolding on Patient class you can see patients have both gender and date of birth, which are in turn of datatypes CD and TS respectively.
Writing some code
So lets write our first GELLO. It simply will be querying for a patients gender.
GELLO can be used in different contexts. Here we will be looking at a single patient at a time. So in the workspace write:
Context SinglePatient
and hit the Compile button  .
.
What happens? Yes there should be a couple of error messages in a new bottom-most pane. The first message is saying it has no idea what you are talking about with the word "SinglePatient"and wants us to specify the model package that can define it. The best way to do this is to get in the habit of adding a line now above the one we have that says:
imports HL7_v2_VMR_V1
Compile that and we are left with the other error message that says in effect , "would you state what it is that you want the code to give you back" (GELLO is declarative). That's ok for now and this error does not stop things from working.
So what we want is the gender of the patient.
Below this put in a blank line and then type:
patient.
and pause - Code autocomplete will give us some hints:
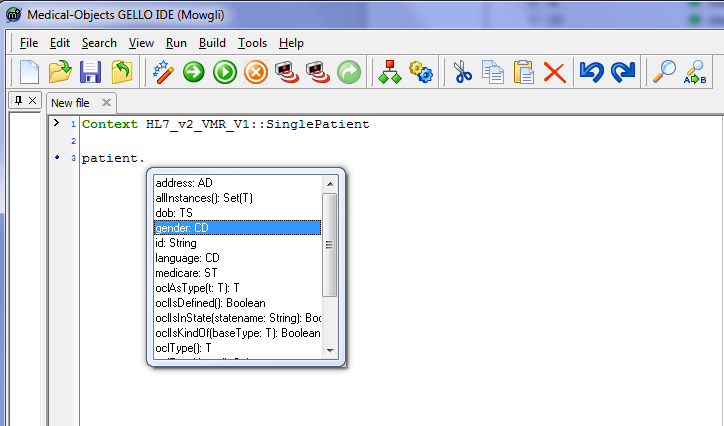
Choose the gender attribute. [Notice anything else about the code in general in this screenshot ? - yes, we can collapse the top two lines into one single line that specifies the imported package and the context.]
Now compile - all good. Ok we choose the Run button  . [ NB - Not the one on the left of this that we can use to step through the code for debugging). The normal procedure is to compile code so that it is syntactically correct, and then run it against some test clinical data.
. [ NB - Not the one on the left of this that we can use to step through the code for debugging). The normal procedure is to compile code so that it is syntactically correct, and then run it against some test clinical data.
Ok, it wants some test data. Here is a small xml file for us to get started with. Cut and past the following text into a text editor such as notepad++ and save as firstTest.xml . Then when we run the GELLO code, navigate to where you have this file.
<?xml version="1.0" encoding="UTF-8"?>
<singlePatient xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:noNamespaceSchemaLocation="./iso-21090-datatypes.xsd">
<patient>
<patientID
root="1.2.36.174030967"
extension="1234567892"
reliability="VRF"
scope="OBJ"/>
<patientName>
<part type="GIV" value="Alice" />
<part type="GIV" value="A." />
<part type="FAM" value="Someone" />
</patientName>
<dob value="19550621"/>
<gender code="248152002"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "Female" />
</gender>
<ssn value="987-65-4320"/>
<address use="WP">
<part type="AL" value="1050 W Wishard Blvd" />
<part type="CTY" value="Indianapolis" />
<part type="STA" value="IN" />
<part type="ZIP" value="46240" />
</address>
</patient>
</singlePatient>
And here is the result - we are looking at top RHS of the editor screen:
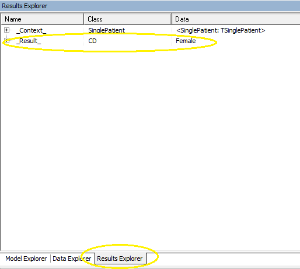
That's it for the first tutorial - well done!
Next time we will write more GELLO code using the date of birth; see how to set some test data in place for a GELLO editing session, and introduce the implies method to check for an allergy.
- No labels
