Often the schema of a clinical domain will be a superset of the VMR. For example, this tutorial considers the situation of a diabetic foot clinic where additional information along with standard observations is recorded in the EHR and presentable through an extended VMR. The VMR extension can be captured in a Package ( in a similar way to the way we did a Library). Its also possible to import the new extended model as a 'hardwired' internal package. Another way to customise an extended data model is to use an archetype.
A diabetic clinic exam will look at the things like the ankle brachial index ( a measure of blood flow), the pulses, and abnormalities that may be present. Compile and then save the following as ExtendedDiabeticFoot.gello_model :
Package ExtendedDiabeticFoot
imports
iso_21090_datatypes,
HL7_v2_VMR_V1
class DiabeticFootObservations extends Any
diabeticFootExamDate: TS
leftDorsalisPedisPulseFull: Boolean
rightDorsalisPedisPulseFull: Boolean
leftPosteriorTibialPulseFull: Boolean
rightPosteriorTibialPulseFull: Boolean
leftAnkleBrachialIndex: Real
rightAnkleBrachialIndex: Real
anyFootDeformity: Boolean
anyNeurologicalChanges: Boolean
anyUlcerPresent: Boolean
class ExtendedDiabeticFootExample_SinglePatient extends SinglePatient
diabeticFootObservations: Sequence(DiabeticFootObservations)
class InfrastructureRoot extends Any
singlePatient: ExtendedDiabeticFootExample_SinglePatient
EndPackage
As before with a Library, this file needs to be saved to the same folder as any GELLO files that use it.
Now lets add some more clinical data to fifthTest.xml and save it as SeventhTuteTest.xml.
Add it between </vitals> and </singlePatient> at the end:
<observations>
<observationCode code="43396009"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "Hemoglobin A1c measurement " />
<translation code="4548-4"
codeSystem="2.16.840.1.113883.6.1"
codeSystemName="LN">
<displayName value = "Hemoglobin A1c/Hemoglobin.Total In Blood" />
</translation>
</observationCode>
<dateTime value = "20160616" />
<value xsi:type = "PQ" value = "9.0" unit = "%"/>
</observations>
<diabeticFootObservations>
<diabeticFootExamDate value = "20160616" />
<leftDorsalisPedisPulseFull value = "false" />
<rightDorsalisPedisPulseFull value = "true" />
<leftPosteriorTibialPulseFull value = "true" />
<rightPosteriorTibialPulseFull value = "true" />
<leftAnkleBrachialIndex value = "0.7" />
<rightAnkleBrachialIndex value = "0.9" />
<anyFootDeformity value = "false" />
<anyNeurologicalChanges value = "false" />
<anyUlcerPresent value = "false" />
</diabeticFootObservations>
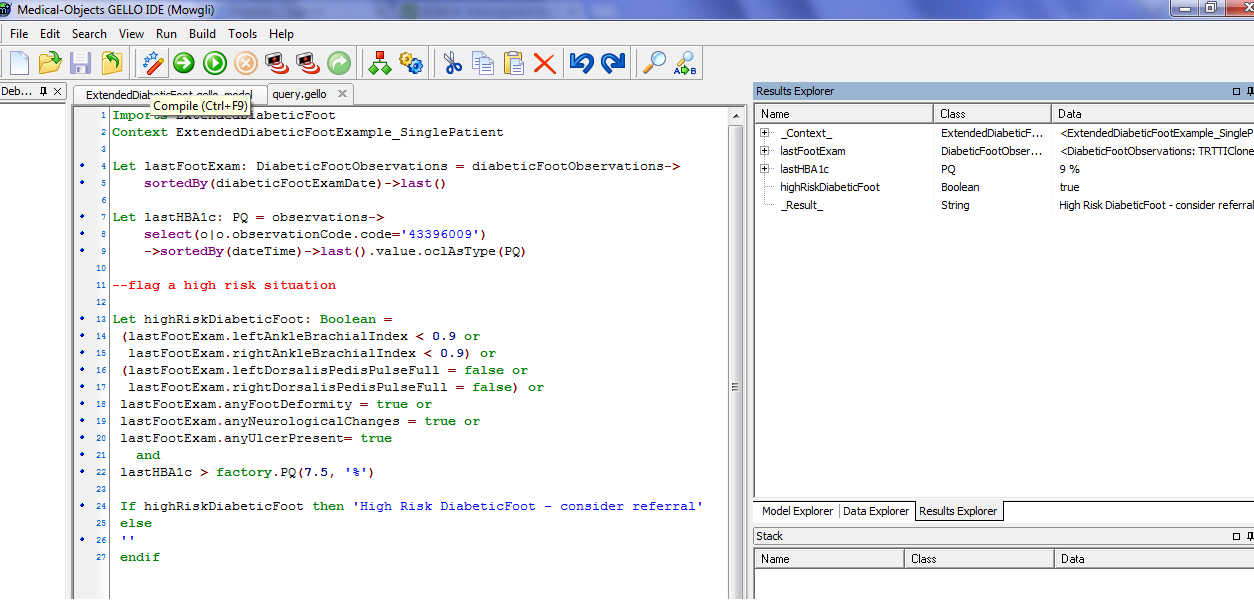
Cut and paste the following GELLO into a new workspace:
Imports ExtendedDiabeticFoot
Context ExtendedDiabeticFootExample_SinglePatient
Let lastFootExam: DiabeticFootObservations = diabeticFootObservations->
sortedBy(diabeticFootExamDate)->last()
Let lastHBA1c: PQ = observations->
select(o|o.observationCode.code='43396009')
->sortedBy(dateTime)->last().value.oclAsType(PQ)
--flag a high risk situation
Let highRiskDiabeticFoot: Boolean =
(lastFootExam.leftAnkleBrachialIndex < 0.9 or
lastFootExam.rightAnkleBrachialIndex < 0.9) or
(lastFootExam.leftDorsalisPedisPulseFull = false or
lastFootExam.rightDorsalisPedisPulseFull = false) or
lastFootExam.anyFootDeformity = true or
lastFootExam.anyNeurologicalChanges = true or
lastFootExam.anyUlcerPresent= true
and
lastHBA1c > factory.PQ(7.5, '%')
If highRiskDiabeticFoot then 'High Risk DiabeticFoot - consider referral'
else
''
endif
And after compiling and saving it say as tuteSeven.gello , run: