GELLO is a functional language. Referential transparency means you can lift an expression from the code and it can substitute for the variable. Loops in a usual sense are incompatible with this paradigm. Loops in GELLO have to be done
with a recursive call, passing the new value as an argument; as a function.
In this worked example for a loop, we will use diagnosis data in both XML and HL7 v2 formats:
XML data
<?xml version="1.0" encoding="UTF-8"?>
<singlePatient xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:noNamespaceSchemaLocation="./iso-21090-datatypes.xsd">
<observations>
<observationCode code="439401001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "Diagnosis" />
</observationCode>
<dateTime value = "20201017" />
<value xsi:type = "CD" code="39579001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "anaphylaxis" />
</value>
</observations>
<observations>
<observationCode code="439401001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "Diagnosis" />
</observationCode>
<dateTime value = "20201017" />
<value xsi:type = "CD" code="195951007"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = " Acute exacerbation of COPD" />
</value>
</observations>
<observations>
<observationCode code="439401001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "Diagnosis" />
</observationCode>
<dateTime value = "20201017" />
<value xsi:type = "CD" code="400047006"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = " Peripheral vascular disease" />
</value>
</observations>
<observations>
<observationCode code="439401001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "Diagnosis" />
</observationCode>
<dateTime value = "20130217" />
<value xsi:type = "CD" code="44054006"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED">
<displayName value = "Diabetes mellitus type II" />
</value>
</observations>
</singlePatient>
Save that as loopObs.xml. Imagine we want to write some GELLO that does a little report on the patient's diagnoses. In this we will loop over the diagnosis observations and extract the diagnosis display names.
The package gello ( ie the file DemoLoopFunctions.gello_model) is:
Package DemoLoopFunctions
imports
iso_21090_datatypes,
HL7_v2_VMR_V1
GetCDDisplayNamesFromSequenceObsAsString_Internal(i:Integer, accum: String, obs: Sequence(Observation)): String =
If i <= obs.size() then
Let intermediate: String
= GetCDDisplayNamesFromSequenceObsAsString_Internal(i+1,
accum + ' ' + obs[i].value.oclAsType(CD).displayName.value + ',' ,
obs
)
Let result: String = intermediate.rtrim(',')
in
result
else
accum
endif
GetCDDisplayNamesFromSequenceObsAsString(obs: Sequence(Observation)): String =
GetCDDisplayNamesFromSequenceObsAsString_Internal(1, '', obs)
EndPackage
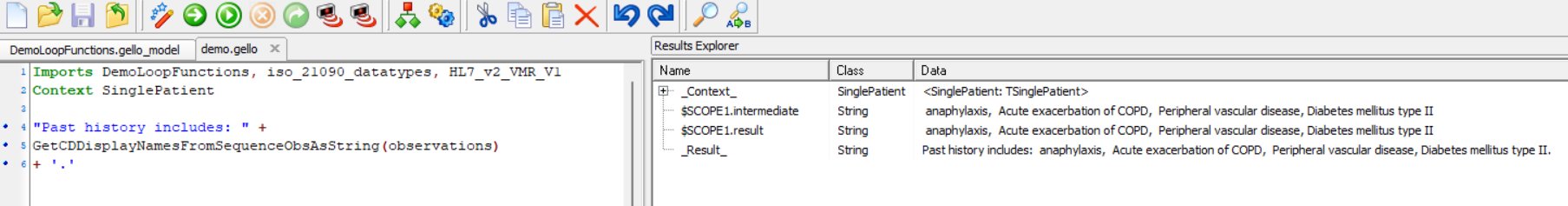
The GELLO that calls this is simply:
Imports DemoLoopFunctions, iso_21090_datatypes, HL7_v2_VMR_V1
Context SinglePatient
"Past history includes: " +
GetCDDisplayNamesFromSequenceObsAsString(observations)
+ '.'
And here is the result:
HL7 v2 data
Here is the same data as an oru file:
MSH|^~\&|MOADLEDIT^MOADLEDIT:3.1.8 [win32-i386]^L|Unassigned^8D9FE669-4710-455D-8B97-811508B616E7^GUID|||20230403184855+1000||ORU^R01|XX04031848133-5109|P|2.3.1^AUS&&ISO^AS4700.2&&L|||||AUS
PID|1
PV1|1|O
ORC|RE||528340EE-4CB1-48DF-8D12-CD8CA891B938^Unassigned^8D9FE669-4710-455D-8B97-811508B616E7^GUID||CM
OBR|1
OBX|1|CE|439401001^Diagnosis^SCT|1.1|39579001^Anaphylaxis^SCT||||||F
OBX|2|CE|439401001^Diagnosis^SCT|1.2|195951007^Acute exacerbation of COPD^SCT||||||F
OBX|3|CE|439401001^Diagnosis^SCT|1.3|400047006^Peripheral vascular disease^SCT||||||F
OBX|4|CE|439401001^Diagnosis^SCT|1.4|44054006^Diabetes mellitus type II^SCT||||||F
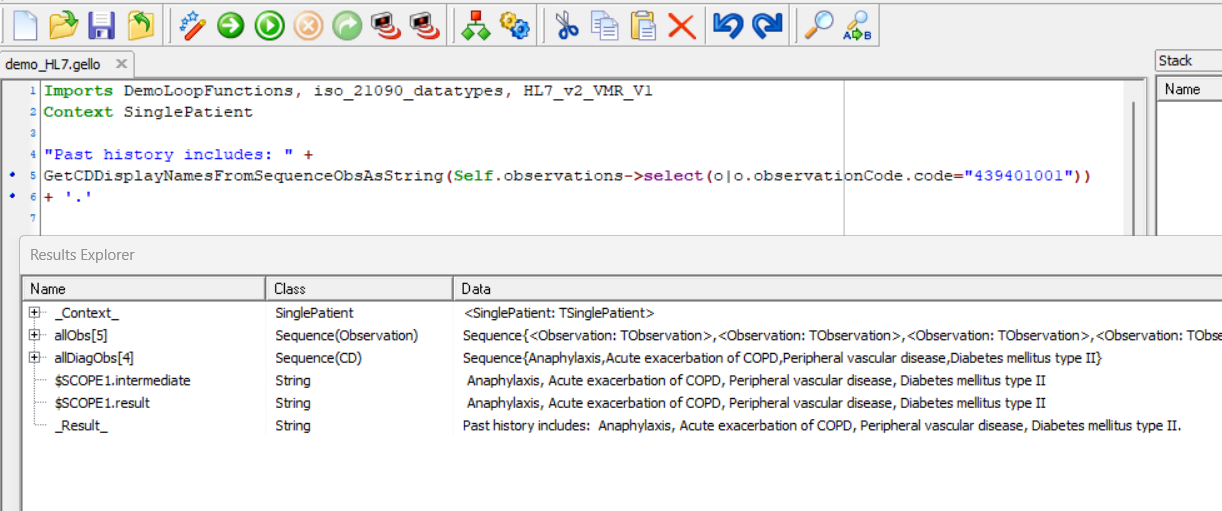
The GELLO for that could become:
Imports DemoLoopFunctions, iso_21090_datatypes, HL7_v2_VMR_V1
Context SinglePatient
"Past history includes: " +
GetCDDisplayNamesFromSequenceObsAsString(Self.observations->select(o|o.observationCode.code="439401001"))
+ '.'
The result is here: