Now we will do an example for checking whether a patient is allergic to penicillin.
Set up a simple GLIF file such as the following, remember to click on the the Allow Modifications button to enable editing:
Give it a name such as Strep_Infn_Check_Penicillin_Allergy.GLIF
Load the patient data file test.xml using the GLIF editor top menu as in the previous tutorial.
Go to the 'Yes' arrow outflowing from the "Allergic to penicillin?" Decision step, and open the GELLO editor for this decision execution.
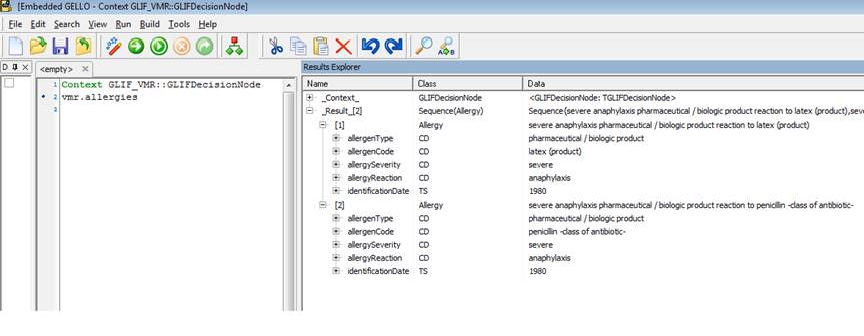
Load up the vmr as before, but this time we are interested in patient allergies , so make the code to be:
Context GLIF_VMR::GLIFDecisionNode
vmr.allergies
Ok we can see two allergies. Unfold on the allergenCode attribute of the second of the two listed allergies in Model Explorer. Note the mandatory three properties of a CD datatype again - the code, codeSystem and codeSystemName. In this case they are '6369005' , '2.16.840.1.113883.6.96' and 'SNOMED-CT'.
Set up the local variable for a parent SNOMED CT concept we want to query on. ( ie this code will need to cover all penicillins in SNOMED CT). Note that in SNOMED CT , medications can be products or substances. Use an online SNOMED CT browser such as that available at http://browser.ihtsdotools.org/ Please note to take care to comply with license restrictions of the IHTSDO available here.
So the code we currently have for the patient is already a high level concept in the Product hierarchy in SNOMED CT. A parent concept for this would be 90614001 | beta-Lactam antibiotic (product) |, but this will subsume cephalosporins as well ( while there is a potential cross allergy to this class, many patients can tolerate them). So we will use the code '6369005', and it just happens to be the same as the code in the patient record in this particular case. l
Let's just use an 'equals' operator for now in the last line:
Let penicillinProductSCT: CD = CD {code = '6369005',
codeSystem = '2.16.840.1.113883.6.1',
codeSystemName ='SNOMED-CT'}vmr.allergies.oclIsDefined() and
vmr.allergies->exists(a | a.allergenCode = penicillinProductSCT)
Here's the result: