Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Overview
This tutorial will build on the GELLO coding introduced last time, with the calculation of an age for our test patient, the adding of clinical data to the test file, the importing of that data into the editor and then looking at the implies method for the first time.
More GELLO writing
Type or cut and past the following code:
| No Format |
|---|
Context HL7_v2_VMR_V1::SinglePatient patient.dob |
Run it and choose the firstTest.xml patient file from the last tutorial:
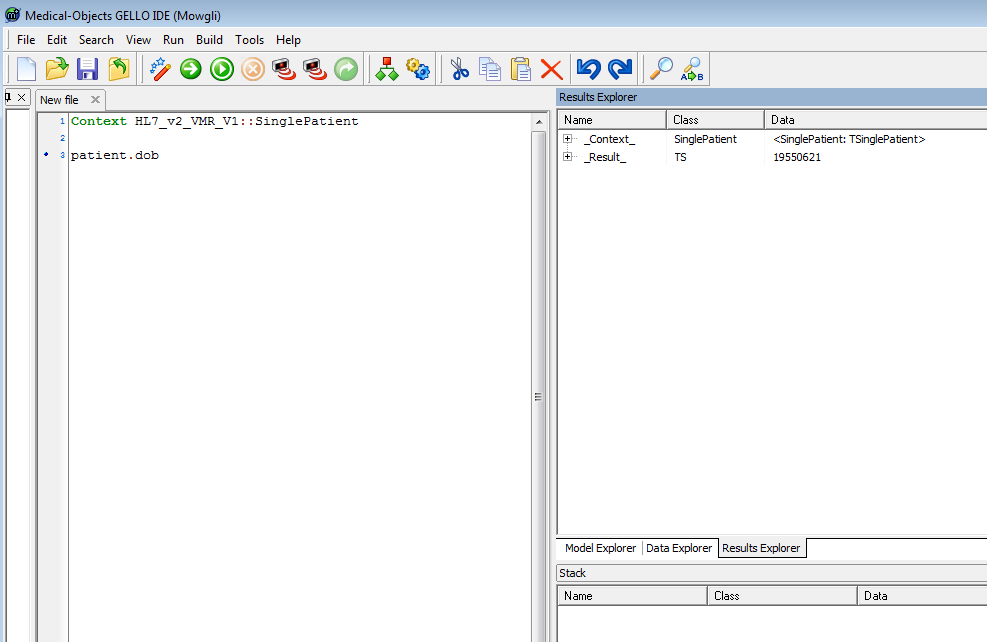
We can see the date of birth is the 21st June 1955, but lets now calculate the patient's age. Assign the retrieved date of birth to be a local variable so that we can calculate with it:
| No Format |
|---|
Let dob = patient.dob |
Compile this and note the error message (note again this will run however). Yes, GELLO is strongly typed and the editor is suggesting the type for the local variable, so add it in. The line becomes:
| No Format |
|---|
Let dob: TS = patient.dob |
We don't worry about the second message saying there is no final expression as we can say that at the end - this returns what it is we actually want - i.e. the age; and we will get to that. So here is where we are up to:
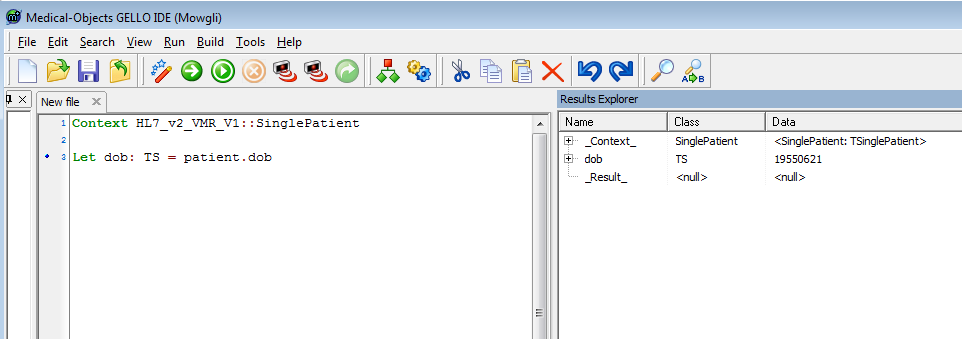
Notice that it says _Result_ is null, again this is because we have no final expression line as yet.
Add in this code which includes the required final expression:
| No Format |
|---|
let ageInSeconds: PQ = factory.TS('today') - dob
Let ageInYrs: PQ = ageInSeconds.convert('yr')
ageInYrs |
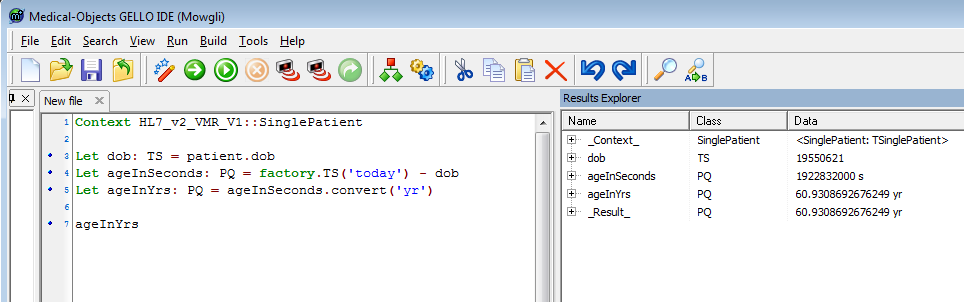
Don't worry too much about the actual code here, but it involves the use of the factory class and a method that converts units.
Unfold on the _Result_ and notice that this Physical Quantity (PQ) typed result has both a unit and a value:
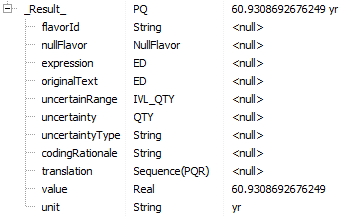
It is best to use PQ variables in GELLO instead of further getting the value out as a Real typed value, eg for laboratory results, as sometimes units change with lab tests and you don't want to be caught out with 'apples and oranges'! [eg think about the two following measurements: 2 cm and 0.7874016 inches. They are the same measurement in fact; but if we just wrote code using the values, 2 does not equate to 0.7874016]
The good thing about using PQs is that you can perform mathematical operations on them. The full list of methods the class understands can be seen in the editor, by looking in the Model Explorer like we did for the CD type (remember GELLO is Object Oriented):
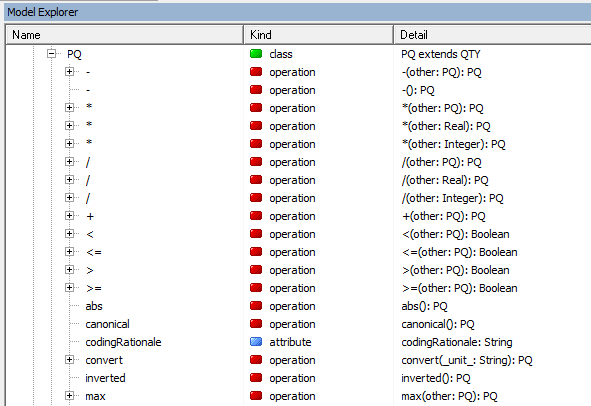
--------------------------------------------------------------------------------------------
Reading Test Data
Ok, next we will move on to how we load up our test data for a GELLO editing session. This is easily done by opening the Model Explorer pane (fold up the PQ and the iso_21090_datatype package if you are still there) and unfolding on the HL7_v2_VMR_V1 package again. Go down to the SinglePatient node and right click on this. Choose the Read Test Data for class option and then choose our firstTest.xml file. This loads up the instance data for the model, which can be viewed in the Data Explorer - so far we have patient class data only.
---------------------------------------------------------------------------------------------
The implies method
Lets add some allergy data to our firstTest.xml file.
Do this by inserting all of this xml above the final line in firstTest.xml and rename it secondTest.xml :
<allergies>
<allergenType code="373873005"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "pharmaceutical / biologic product" />
</allergenType>
<allergenCode code="111088007"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "latex (product)" />
</allergenCode>
<allergySeverity code="24484000"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "severe" />
</allergySeverity>
<allergyReaction code="39579001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "anaphylaxis" />
</allergyReaction>
<identificationDate value="1980"/>
</allergies>
<allergies>
<allergenType code="373873005"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "pharmaceutical / biologic product" />
</allergenType>
<allergenCode code="6369005"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "penicillin -class of antibiotic-" />
</allergenCode>
<allergySeverity code="24484000"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "severe" />
</allergySeverity>
<allergyReaction code="39579001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "anaphylaxis" />
</allergyReaction>
<identificationDate value="1980"/>
</allergies>
This data accords with the structure of the allergies class in our VMR.
Now lets move to the editor and think about how we will look for a penicillin allergy.
We will use the implies method. This works for data that is of a CD data type. We can see from the above data that the allergenCode-tagged element has several attributes - namely code, codeSystem and codeSystemName. These three are all required by the ISO21090 standard to make a CD.
So lets load up our new secondTest.xml file into the editor as before by right clicking on the SinglePatient node in the HL7_v2_VMR_V1 package in Model Explorer and bring it in with Read Test Data for class. Notice the new allergy data in Data Explorer view. Make sure all attributes are visible and not null. (If things are null there has usually been an error in the syntax of the xml)
Now to the workspace, let's test we can access this data:
| No Format |
|---|
Context HL7_v2_VMR_V1::SinglePatient Let patientAllergies: Sequence(Allergy) = allergies |
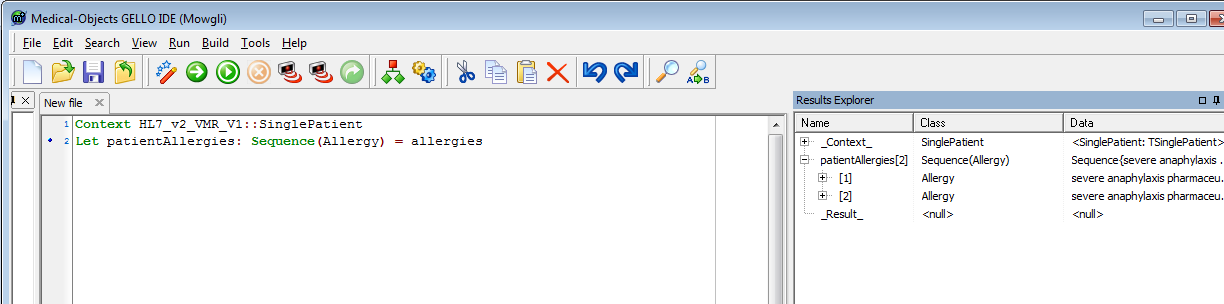
We now create a local or temporary variable which will act as parent concept for subsumption checking in SNOMED CT, using the implies method.
| Info |
|---|
PLEASE NOTE: Licensing restrictions apply to the use of SNOMED CT , please see ihtsdo.org for further information. In general if you are a citizen of a country that is a member of the IHTSDO usage is free, but still requires a license.] |
add this:
| No Format |
|---|
Let penicillinClass_SCT:CD = CD{code = '6369005',
codeSystem = '2.16.840.1.113883.6.96',
codeSystemName ='SNOMED-CT'
} |
So that's the parent concept - the class of penicillins. Our test data happens to be at this general level as well - implies works as "equals" as well as "is a child of". The implies method is a collection operator, and allergies from the VMR is a sequence; so we use the following syntax and grammar - add:
| No Format |
|---|
Let hasPenicillinAllergy: Boolean = allergies->exists(a| a.allergenCode.implies(penicillinClass_SCT).asBoolean()) hasPenicillinAllergy |
Run it:
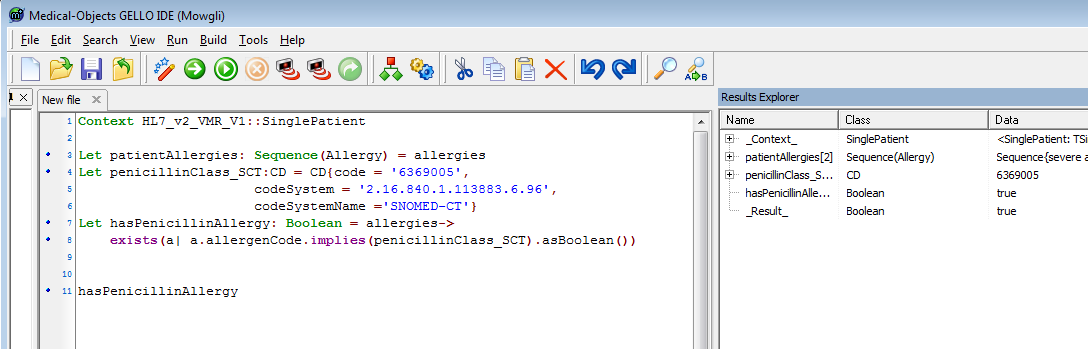
We can shorten the code needed to create the parent class local variable - by replacing line 4 with:
| No Format |
|---|
Let penicillinClass_SCT: CD = factory.CD_SNOMED_CT('6369005')
|
OK, lets now see what happens if instead of saying the patient is allergic to all penicillins they have been noted to be allergic to a member of that class, in the EHR; for example amoxicillin ( - normally this would mean they are also allergic to all penicillins):
So we change the data in the xml:
<allergies>
<allergenType code="373873005"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "pharmaceutical / biologic product" />
</allergenType>
<allergenCode code="27658006"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "Amoxicillin" />
</allergenCode>
<allergySeverity code="24484000"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "severe" />
</allergySeverity>
<allergyReaction code="39579001"
codeSystem="2.16.840.1.113883.6.96"
codeSystemName="SNOMED-CT">
<displayName value = "anaphylaxis" />
</allergyReaction>
<identificationDate value="1980"/>
</allergies>
Save this as secondTest_amoxicillin_allergy.xml, and run it this time with this data using the Remote execute with xml patient data button:
You should get a true result when looking in the Results Explorer in spite of any error messages:
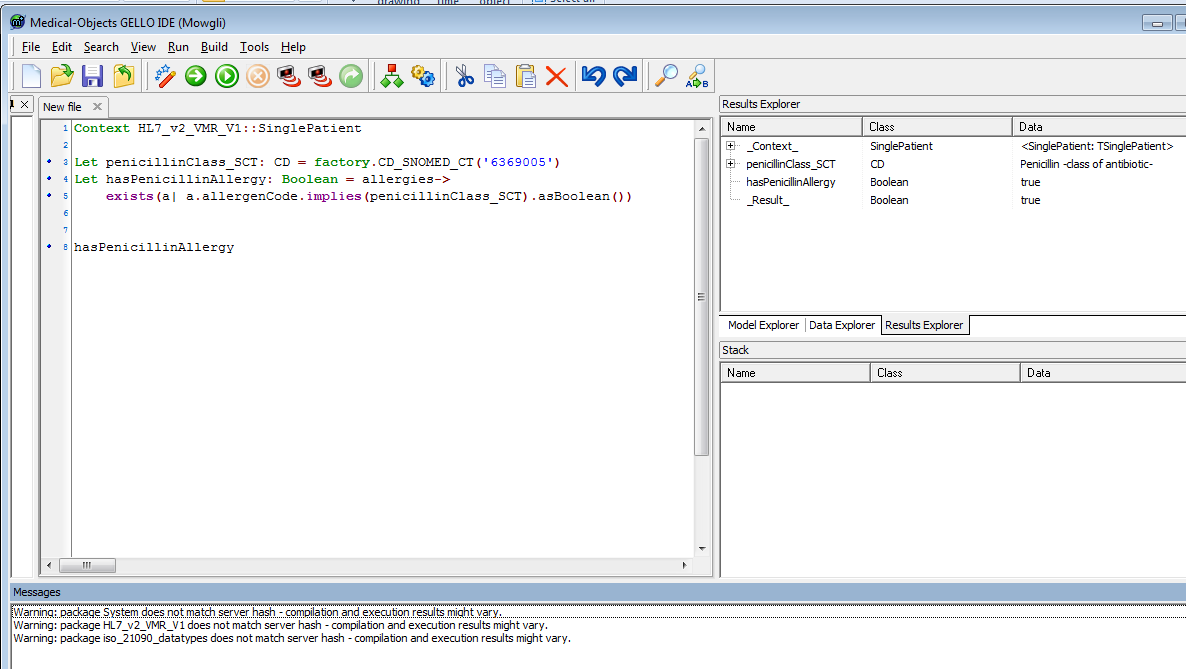
This demonstrates the real value of a reference terminology such as SNOMED CT. If we were using ICD - 9 for example we would have to input a set of codes for all penicillin antibiotics, here we just use implies to see if our allergen is a penicillin - simple as that.
That completes this tutorial.