Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
This worked example will walk you through the insertion of GELLO code on decision arms for the '? anaemia' decision in the GORD (Gastro-oesophageal reflux) example. A GLIF file is provided (GORD_start.GLIF) that is suitable to use; along with some test patient data in xml format.
Anaemia is a low haemoglobin (hemoglobin) level, and in the GORD guideline example represents a compelling reason for early endoscopy, as there may be a cancer of the esophagus or stomach causing the reflux symptom.
Open and unzip the attached folder. Open the GLIF file with the GLIF Editor.
It opens at the "top level" guideline. There are three sub-guidelines and they can be accessed by clicking through the guideline itself or by scrolling down on right hand side (RHS) of the menu bar:
Go to the "consider early endoscopy" sub-guideline.
Notice the decisions, as elsewhere are pink. They are not automated as yet. So we will do that for the anaemia decision.
You can write GELLO for GLIF decisions and test it against loaded patient data. Let's load some data. This is in xml format and you can look at the test data in notepad++ as well if you like.
Go back to the top level guideline then go File, Load xml Test data. Choose test.xml. Go back to the "consider early endoscopy" guideline.
Now click on the Allow modifications button ![]() ( count six in from the LHS of the top menu.)
( count six in from the LHS of the top menu.)
Position the cursor on the "yes" arrow coming out of the "Anaemia?" decision such that you get a little hand with the finger pointing onto the arrow. Double click and go Edit:
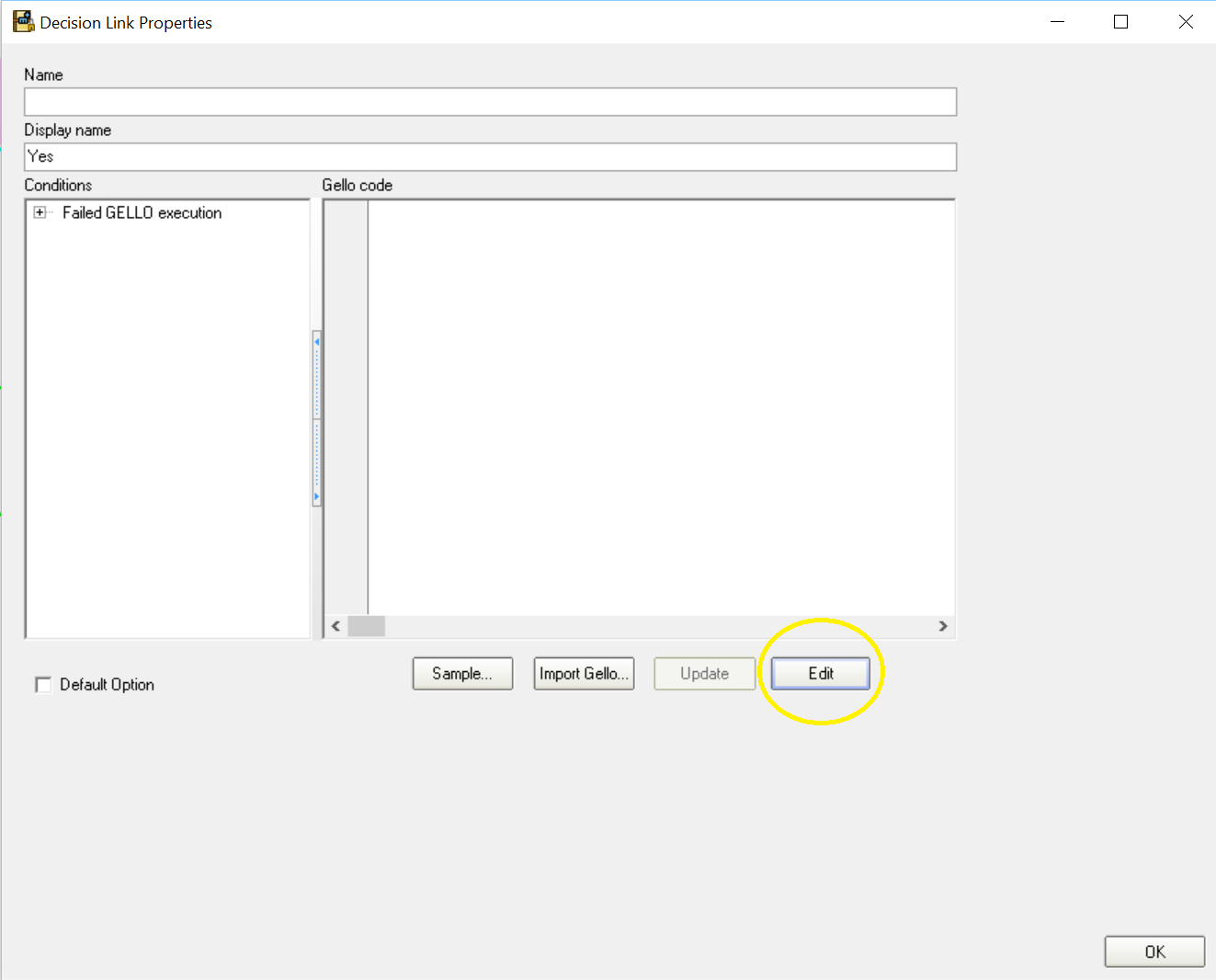
.. to get to the GELLO editor screen:
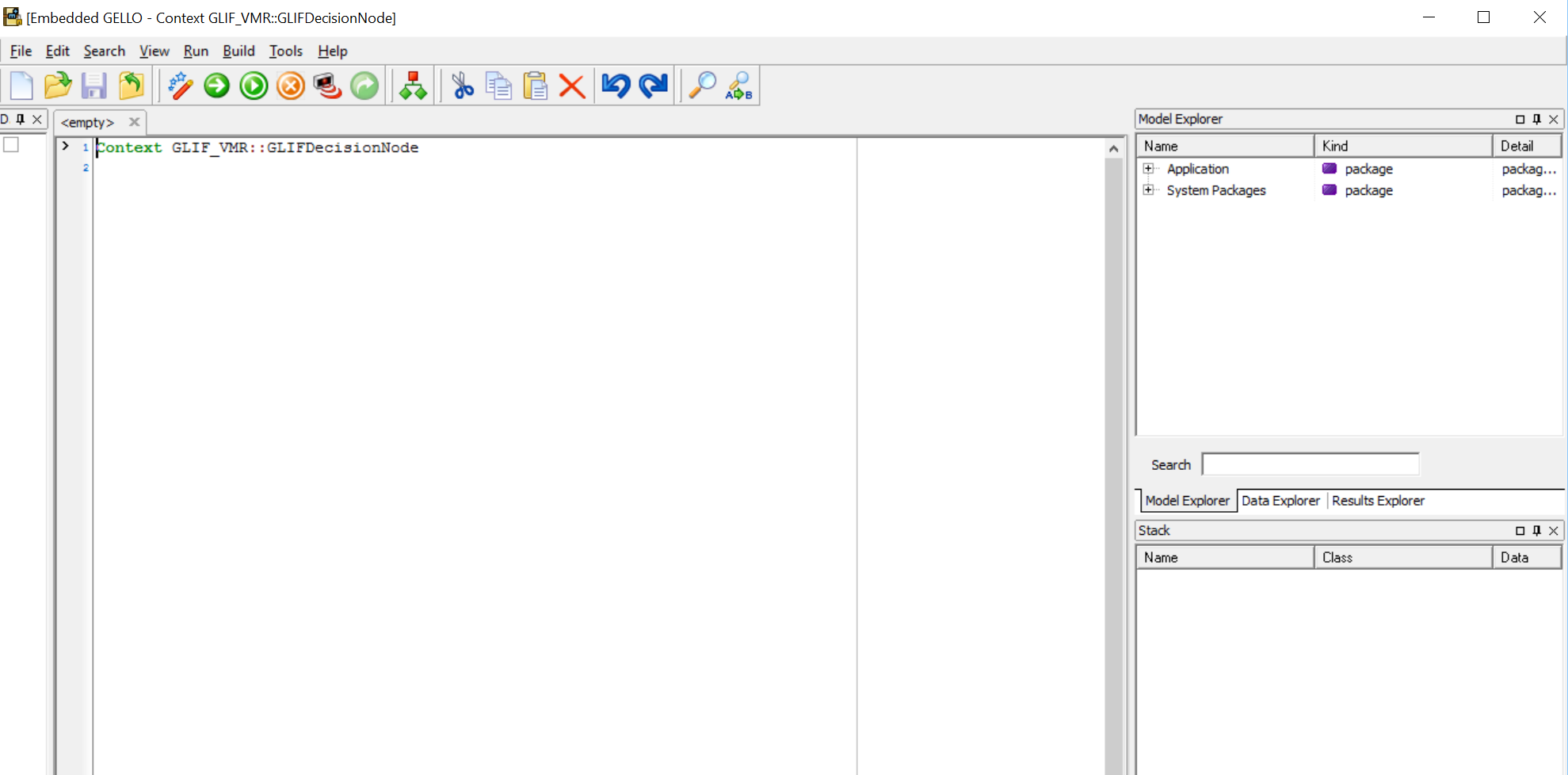
The main panes are the middle workspace and the RHS Model Explorer, Data Explorer and Results Explorer.
Lets take a minute to look at the Model Explorer.
Unfold on System Packages. From the bottom up, GLIF_VMR models a representation of GLIF that accords with 'level B' described on page 7 of the document GLIF_TECH_SPEC_May_4_2004.pdf. So this is the model that understands where and what we can see and do with GELLO in Level A GLIF artifacts such as nodes/steps. Make particular note of the attributes required for a GLIFDecision Result as we use them later on. Above GLIF_VMR is HL7_v2_VMR_V1. This is a draft HL7 standard virtual medical record model that Medical Objects balloted successfully in 2013. Most of the world uses V2 messaging so we can build a CDS suitable model out of what people currently use and send back and forth.
Notice some classes in there such as Allergy and Observation. These classes have attributes that use ISO 21090 data types such as CD and TS:
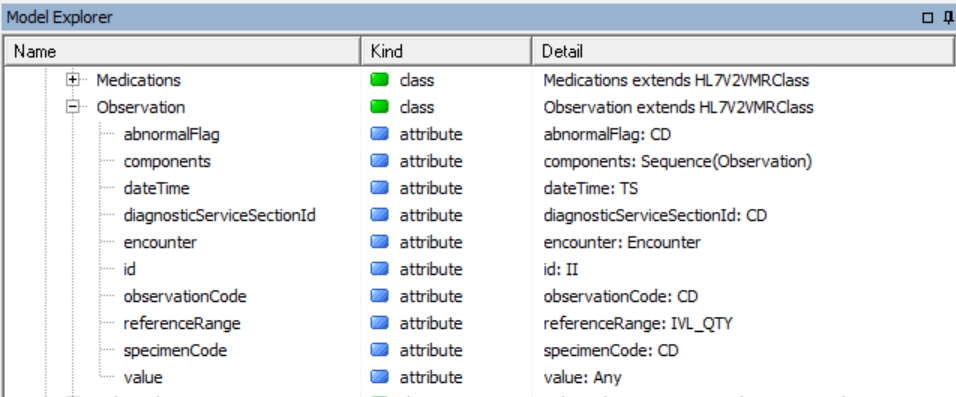
Fold all that up and the next level up is SNOMED CT. SNOMED CT is a large medical ontology looked after by IHTSDO.org. GELLO uses it especially for subsumption queries, with the "implies"method. We won't see this today but it is useful in the homework at the end ;-)
| Info | ||
|---|---|---|
| ||
| PLEASE NOTE: Licensing restrictions apply to the use of SNOMED CT , please see ihtsdo.org for further information. In general if you are a citizen of a country that is a member of the IHTSDO usage is free, but still requires a license.] |
Above this is iso_21090_datatypes. If you look up the CD datatype in here you can see that is about coded values and that the implies method is understood by this datatype.
Ok lets go back to the middle workspace. Notice the editor puts the first line in for you. We are in a context of a GLIF Decision node.
Our first line of GELLO below this context statement will be:
1 + 1
Compile it (five buttons in from LHS top menu). All good, now run it (seven buttons in from LHS top menu). Can you see the result now in Results Explorer?
Because we have loaded test data we can see it and start to think about how we can get to the data we need for a decision as to whether the latest hemoglobin result (if it exists) is too low.
To just see what we have loaded, clear the '1 + 1' line and simply type 'vmr'
Run that and unfold the result in Results Explorer. Can you see 31 Observations? Have a look at the first one:
Class Observation gets used a lot. Here we can see it has an observationCode (unfold that too for a look - see it in turn has a code, a codeSystem and a codeSystemName); a value which is of type Physical Quantity and a dateTime.
Returning to the workspace code, when using the stand alone GELLO editor we can specify what packages are being imported. This is useful for an extended vmr model or to import a Library with its pre-made functions.
Saying 'Imports HL7_v2_VMR_V1 ' directly after the Context statement is important in other contexts, but is usually not needed in the GLIFEditor environment.
Lets get all the Hemoglobin observations.
We can see in the image from just above that observations is a sequence of instances of class Observation. So we can use a select operator as this is a sequence and make use of the observationCode.
Here's two lines to put in now in the workspace. We will make local variables with 'Let' statements. Clear the 'vmr' line we had and cut and paste:
Let hemoglobin:CD = CD{code = '718-7',
codeSystem = '2.16.840.1.113883.6.1',
codeSystemName ='LN'}
Let allHbObservations = vmr.observations->select(observationCode = hemoglobin)Compile and run. The first line makes a local variable of type CD that is then plugged in the select operator in the second. The codeSystemName is 'LN" which is short for LOINC and 718-7 is the code. [Find this in the text.xml data if you like. You can read about this code by googling "hipaa 718-7". Loinc is produced by the Regenstrief Institute.]
Now are there any error messages? - look down the bottom... Unlike 'hemoglobin:CD' we haven't specified a datatype for the variable 'allHbObservations'. This compile error didn't stop the code we have running, so its not critical. It is in fact sometimes useful to leave a LHS variable untyped and to see what is actually being returned from the RHS; but let's put in what is now suggested, and the line in question becomes:
Let allHbObservations: Sequence(Observation) = vmr.observations->select(observationCode = hemoglobin)
Note vmr.observations is a sequence of Observation
The second error message is that there is no final expression. Gello is made of an inner expression (in) an outer expression. We haven't done the final declarative line yet that constitutes the outer expression. This will be the GLIFDecisionResult object we come up with at the end of this GELLO expression.
Making use of the populated dateTime attribute in class Observation:
Let mostRecentHb = allHbObservations->last()
If you run/compile that couple of times you should again get the error suggesting a type for LHS, so becomes:
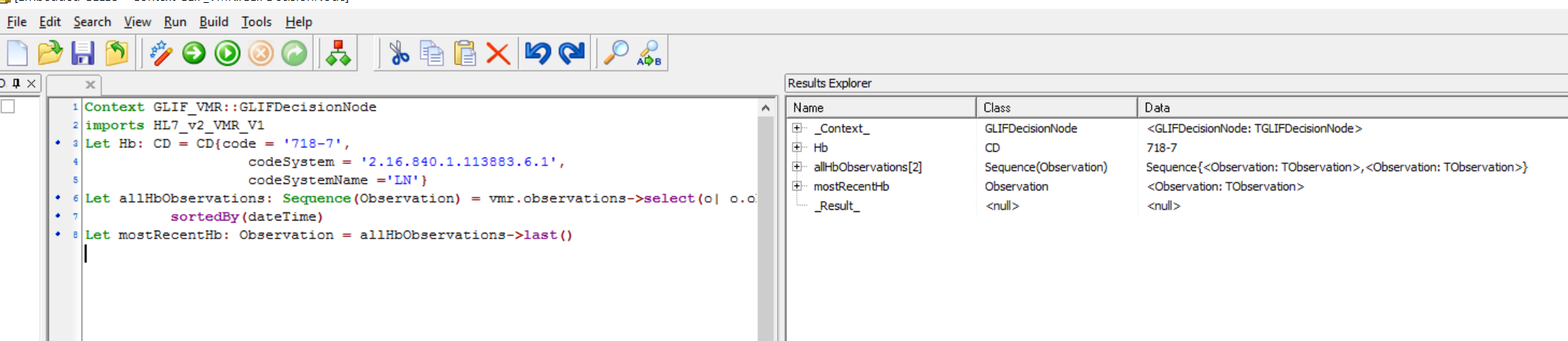
Let mostRecentHb: Observation = allHbObservations->sortedBy(dateTime)->last()
and screenshot after running is :
Now we need to see if mostRecentHb is too low ( ie if the patient is anemic). Make a threshold for anaemia, say 115 g/l.
Let anaemiaThreshold: PQ = factory.PQ(115,'g/L')
Yup - there is a factory to which we send a method PQ (to make a Physical quantity - which has attributes of value: Real and units: String)
Need to make 'mostRecentHb' a PQ as well (currently an Observation); so
Let mostRecentHBAsPQ: PQ = mostRecentHb.value.oclAsType(PQ)
We are getting the value of the Observation ( which here is a PQ type) but we need to make it explicitly a PQ using the oclAsType(PQ) method. Its a GELLO 2 thing...
[As an aside when you have just put a dot after some code, there is code autocomplete available which can help guide you ]
Now we compare the two to get a boolean intermediate answer:
Let hemoglobinIsLow:Boolean = mostRecentHBAsPQ < anaemiaThreshold
and running this returns true for the 'hemoglobinIsLow' variable.
But we need a final declarative statement, and this should be a type of GLIFDecisionResult as we are in a GLIF decision arrow for "yes" , so the final GELLO to be added is:
--need to return a GLIFDecisionResult as we are doing a GLIF Decision
let q: String = "Is hemoglobinhaemoglobin below 115 g/l?"
let aWeight: Integer = 50
Let result:GLIFDecisionResult =
if not mostRecentHb.oclIsDefined() then
GLIFDecisionResult{Question = q,Answer = unknown, Reason = "No haemoglobin observations",Weight = aWeight}
else
if hemoglobinIsLow
then
GLIFDecisionResult{Question = q,Answer = true, Reason = "Haemoglobin is above threshold",Weight = aWeight}
else
GLIFDecisionResult{Question = q,Answer = false, Reason = "Haemoglobin in not above threshold",Weight = aWeight}
endif
endif
result
Ok here we have two new variables for the result object ( as required for a GLIFDEcisionResult - see the model in model explorer), a test for no data, and if-then statements for the final result.
Here is screen shot of this working in the GLIFeditor's gello editor we are using:
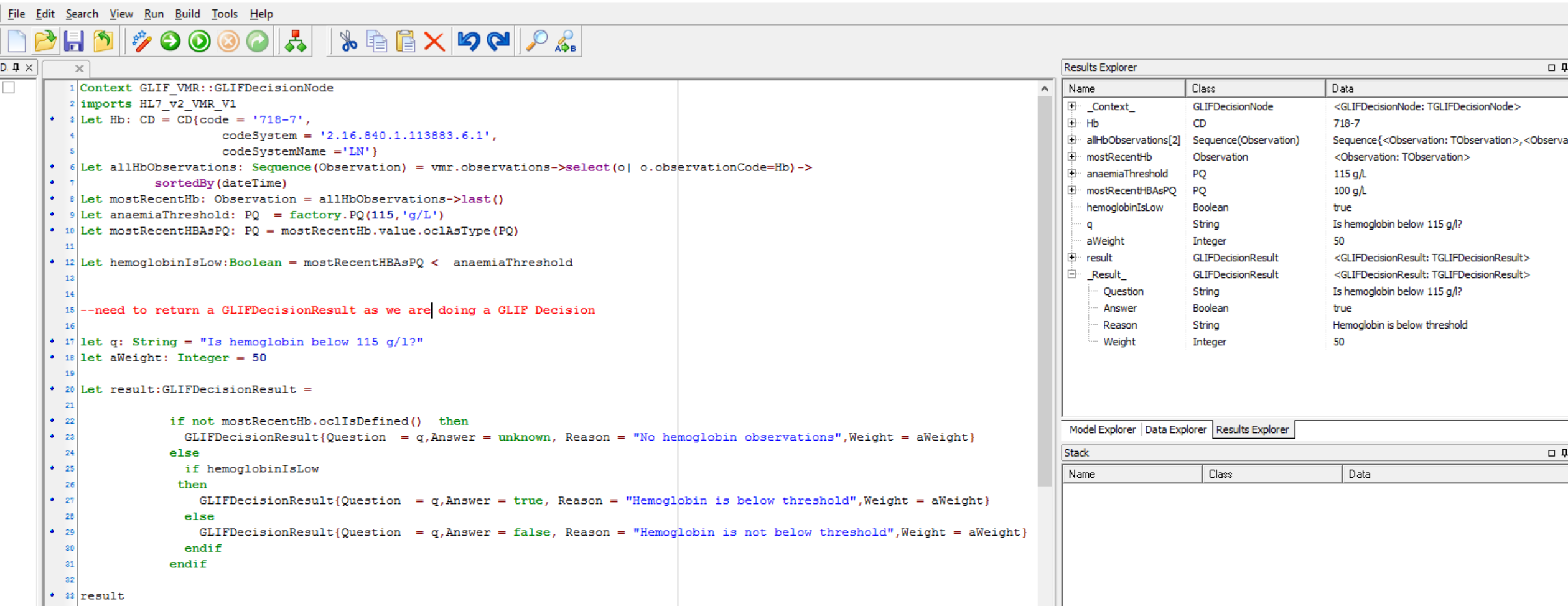
Not quite finished. Its a good idea to put GELLO in all decision arrows coming out of an automated GLIF Decision. So for "no" we would be very similar to this but just change around the final result.
This is how it looks now after clicking through the first pink colored decision:
You can save and reuse the state of this GLIF by File, Save State; then re open the GLIF file as a new instance in the editor and go File, Run Glif with State.
Maybe you can try building a new simple GLIF file now as 'homework', for a decision as to whether a patient (test.xml) has a penicillin allergy .
This GLIF file is not yet complete. Another exercise we could do is to look at the Decisions in management subguideline, adding GELLO to make the decisions automatic but using persisted, previously entered archetype data from the top level. Using ISO 13606 archetypes in GLIF will be another tutorial topic.